发表论文
-
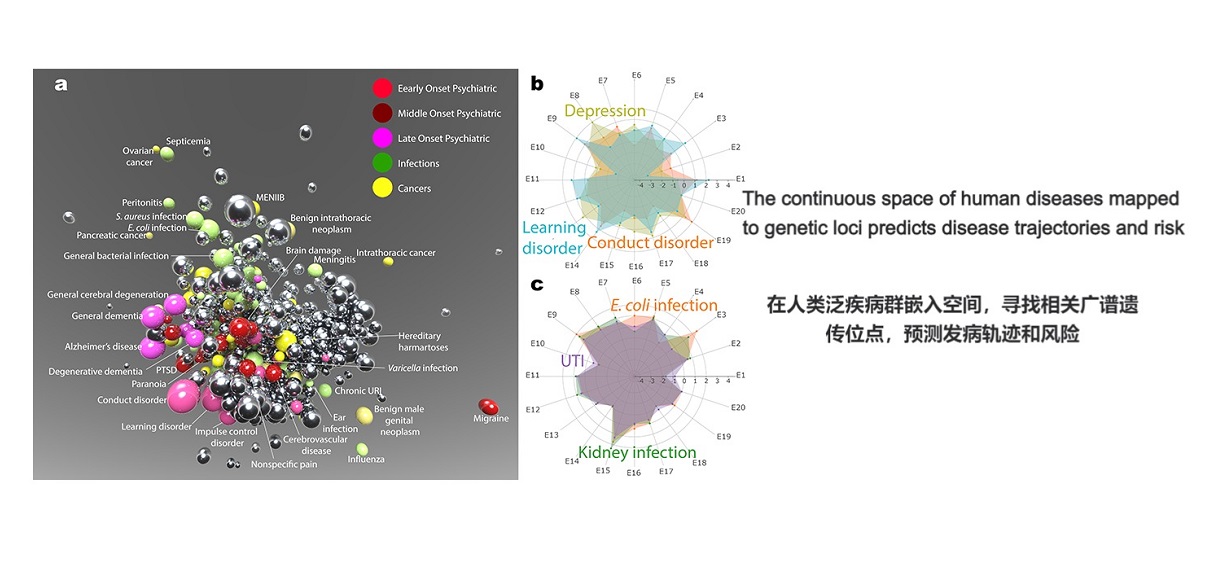
. G. Jia#*, Y. Li#, …, X. Gao*, A. Rzhetsky*. The high-dimensional space of human diseases built from diagnosis records and mapped to genetic loci. Nature Computational Science (2023).(#co-first, *co-corresponding)
-
. Chen chen#, …, G. Jia*, Ta-na Wuyun*, Construction of Apricot Variety Search Engine Based on Deep Learning. Horticultural Plant Journal (2023).
-
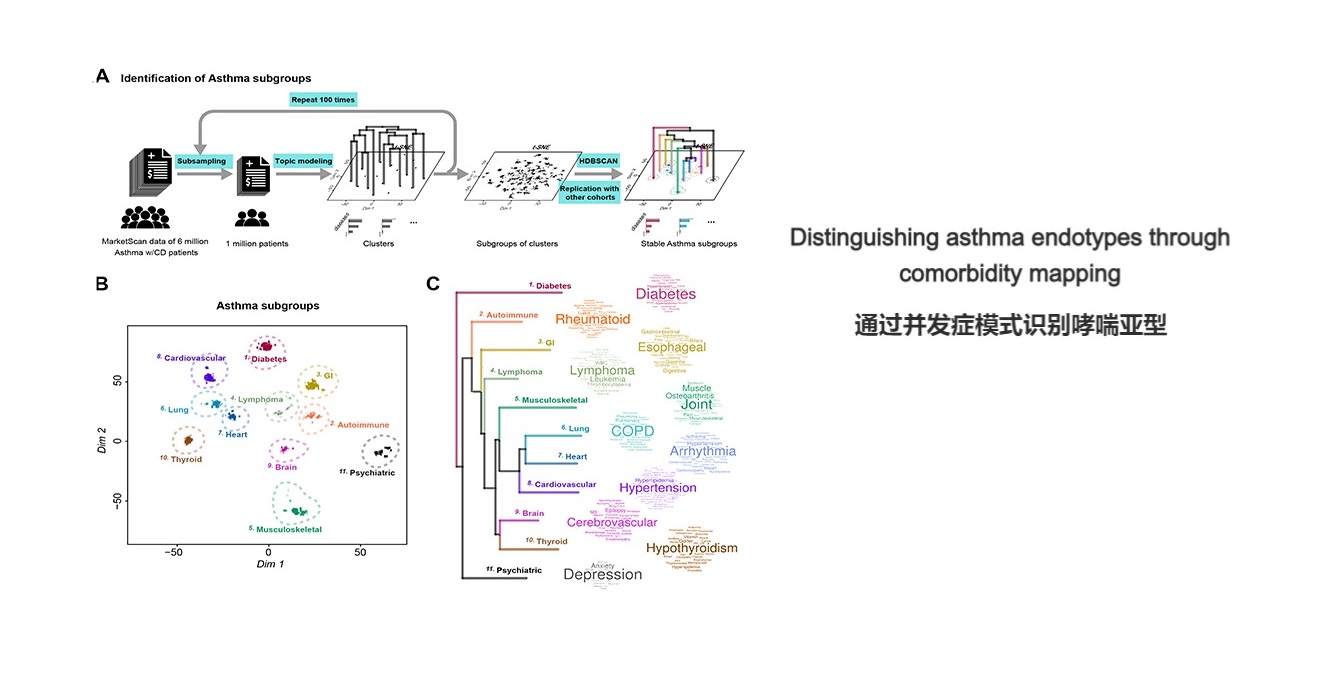
. G. Jia, Zhong, X., Im, H.K. et al. Discerning asthma endotypes through comorbidity mapping. Nature Communications (2022).
-
. X. Zhong, Z. Yin, G. Jia, D. Zhou, Q. Wei, A. Faucon, P. Evans, E. R. Gamazon, B. Li, R. Tao, A. Rzhetsky, L. Bastarache, N. J. Cox, Electronic health record phenotypes associated with genetically regulated expression of CFTR and application to cystic fibrosis. Genetics in Medicine (2020).
-
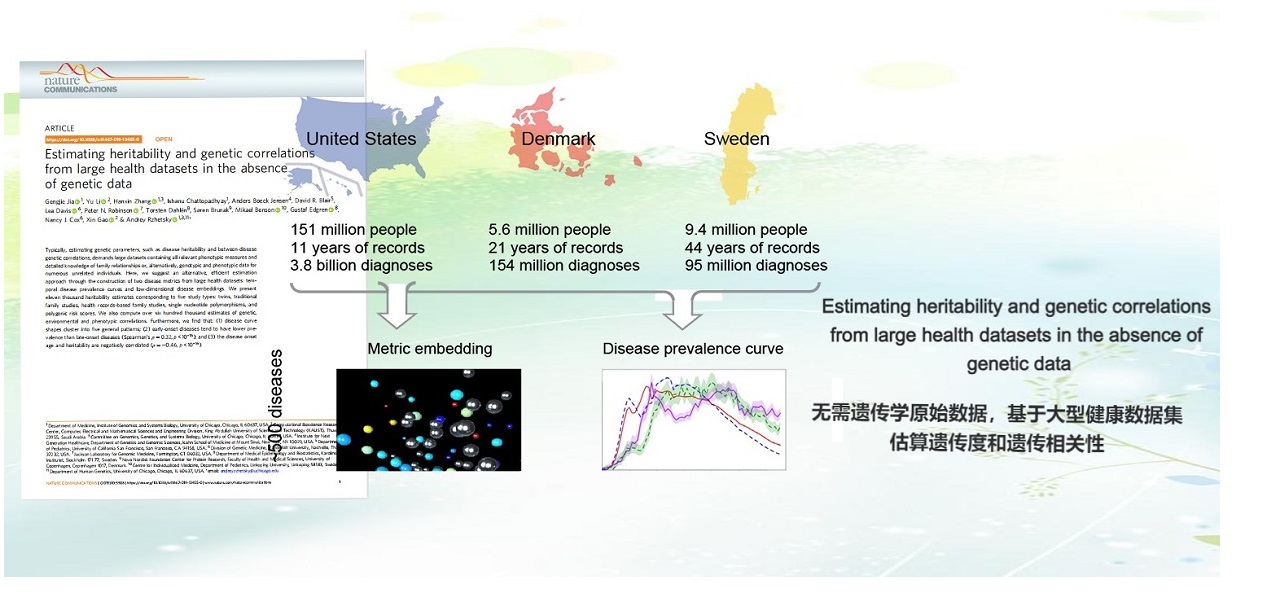
. G. Jia, Y. Li, H. Zhang, I. Chattopadhyay, A. B. Jensen, D. R. Blair, L. Davis, P. N. Robinson, T. Dahlén, S. Brunak, M. Benson, G. Edgren, N. J. Cox, X. Gao, A. Rzhetsky, Estimating Heritability and Genetic Correlations from Large Health Datasets in the Absence of Genetic Data. Nature Communications (2019).AAAS news highlight | Sex-and-country-stratified prevalence curves for over 500 diseases | Machine learning features for heritability and correlation imputation
-
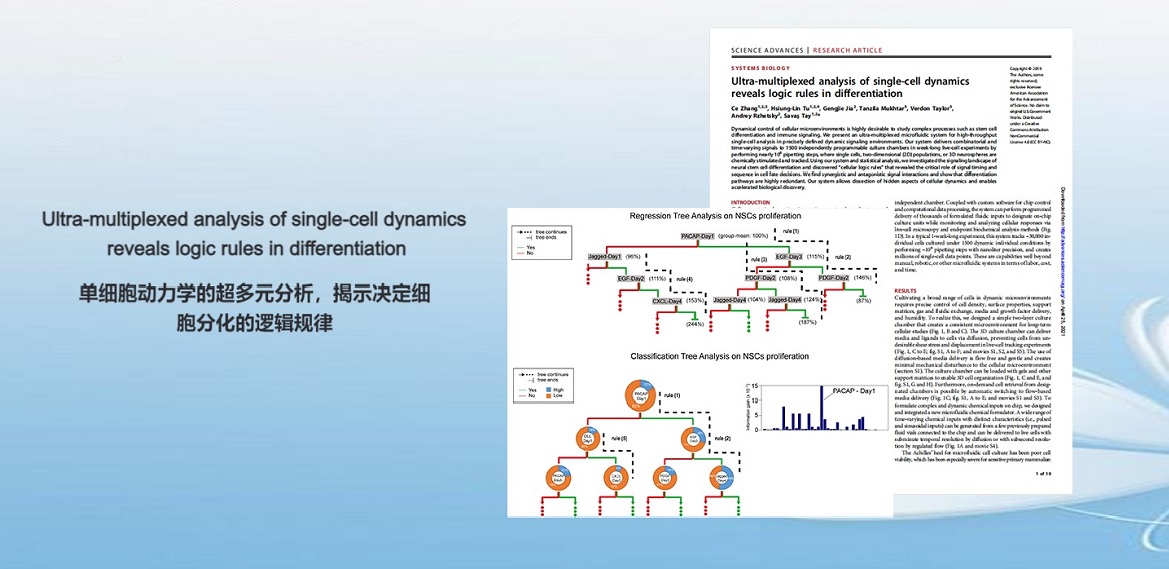
. C. Zhang, H. Tu, G. Jia, T. Mukhtar, V. Taylor, A. Rzhetsky, S. Tay, Ultra-multiplexed Analysis of Single-cell Dynamics Reveals Logic Rules in Differentiation. Science Advances (2019).
-
. G. Jia, G. Stephanopoulos, R. Gunawan, Ensemble Kinetic Modeling of Metabolic Networks from Dynamic Metabolic Profiles. Metabolites (2012).
-
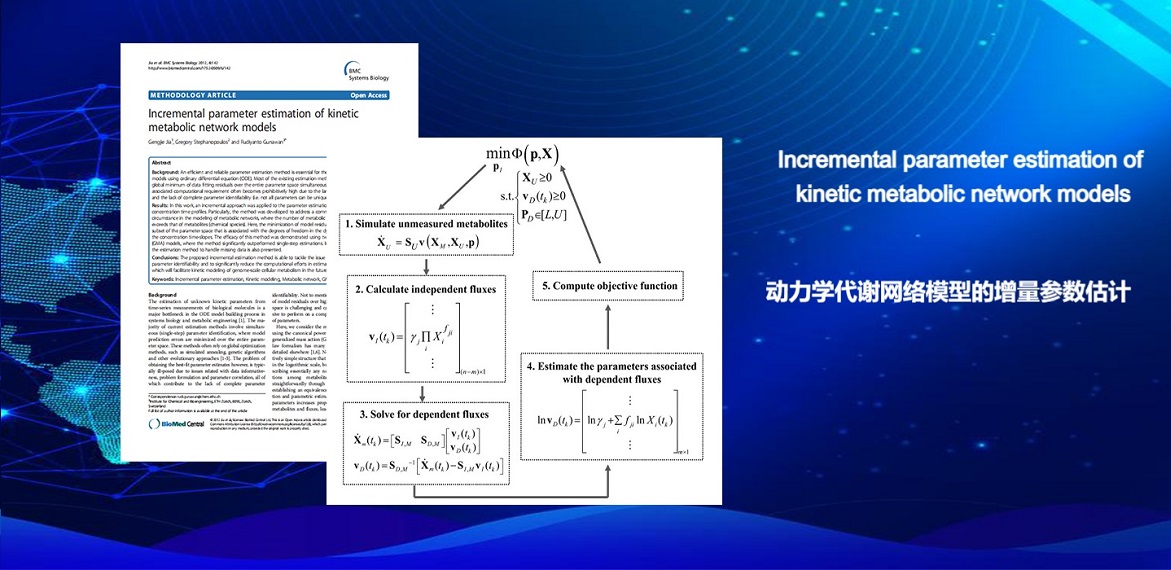
. G. Jia, G. Stephanopoulos, R. Gunawan, Incremental Parameter Estimation of Kinetic Metabolic Network Models. BMC Systems Biology(2012).
-
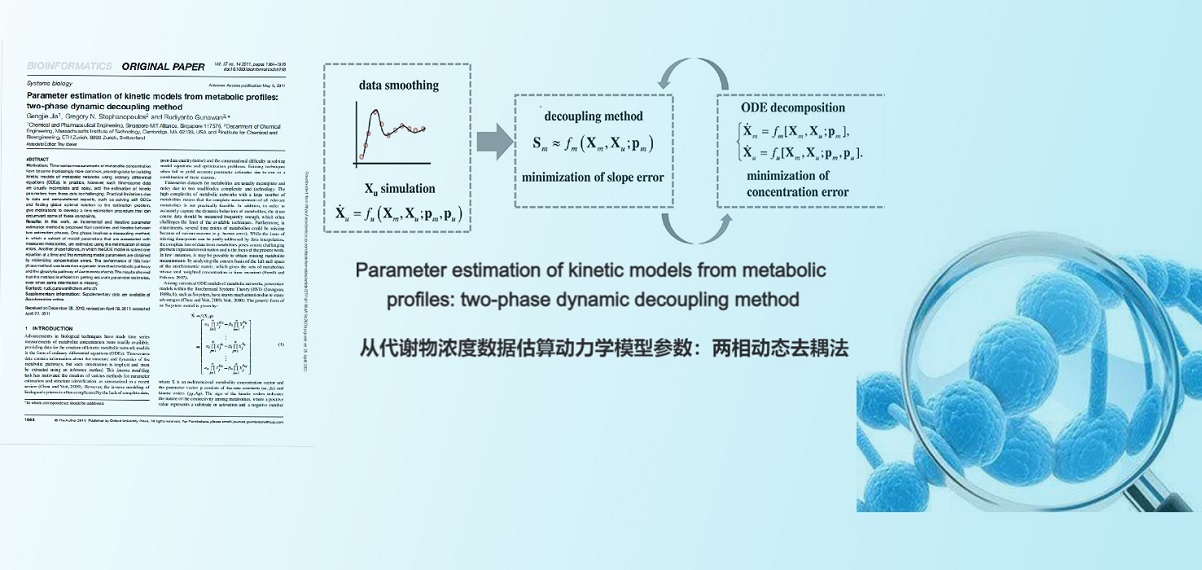
. G. Jia, G. Stephanopoulos, R. Gunawan, Parameter Estimation of Kinetic Models from Metabolic Profiles: Two-phase Dynamic Decoupling Method. Bioinformatics(2011).